CloneAssist
The CloneAssist add-on offers a molecular cloning suite to support Molecular Biology R&D labs. With CloneAssist you can visualize DNA and plasmids and apply various molecular cloning techniques.
CloneAssist is released as beta version and may contain bugs that create irregular behavior/glitches in the software or generate unexpected or incorrect results. By installing CloneAssist you confirm that you understand the use of this add-on may cause irregularities and that eLabNext cannot be held responsible for any damage that this may cause. During this phase we offer a free extended 1-year trial. Please contact us for more information on how to apply for the extended free trial.
To use CloneAssist, go to the Marketplace and install the CloneAssist add-on.

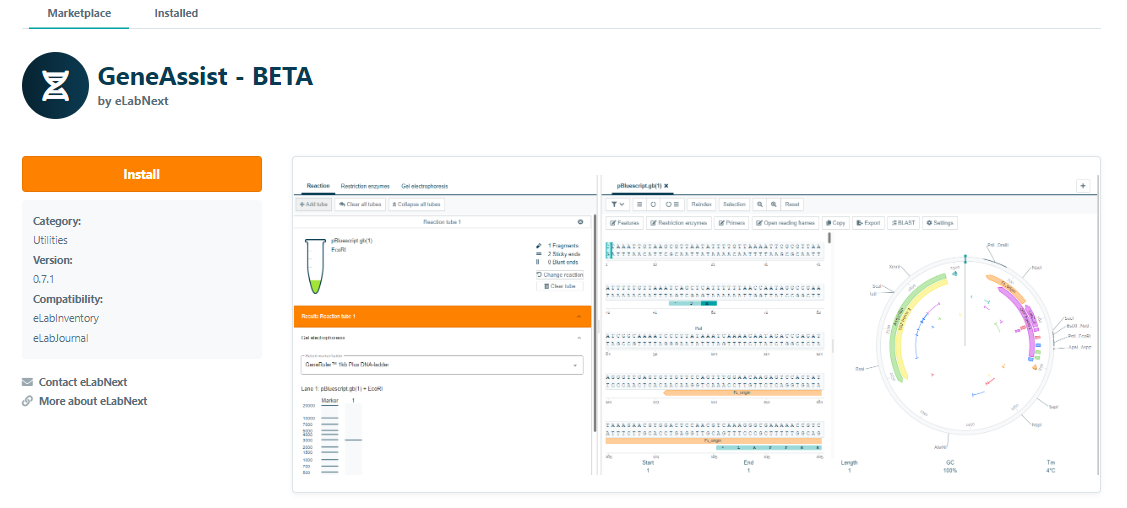
Once the add-on has been installed, CloneAssist is available as a new section type to add to the ELN.
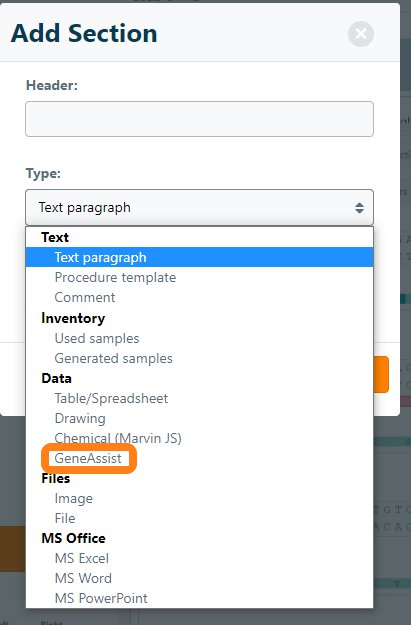
After creating the section, you can start a new CloneAssist project by clicking Start new.

In CloneAssist you can visualize DNA molecules and the corresponding characteristics, annotate DNA molecules, perform digestion reactions, perform ligation reactions, and perform PCR reactions.
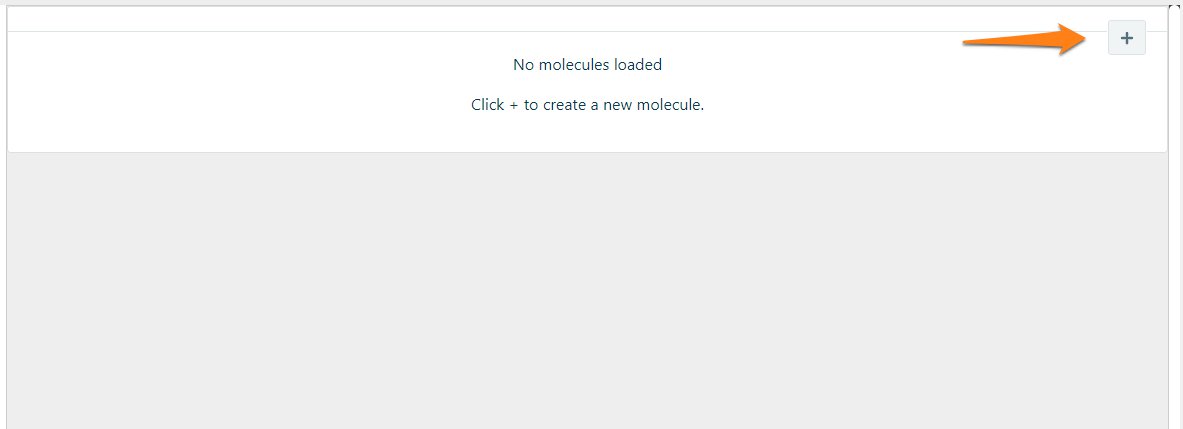
To add new molecules to CloneAssist Click on + and load new molecules. Supported formats are GenBank (.gb, .gbk), FASTA (.fasta, .fa), SnapGene (.dna) or CloneManager (.cm5) files.
Save your CloneAssist section by clicking on Save. Close the section by clicking on Close.

Visualize and annotate DNA molecules
Visualize your molecules in either a linear, circular, or combined view. The important features, restriction enzymes, and open reading frames are directly visualized. To retrieve information of the feature, click on the feature and get the type, start and end-position, length, strand, GC content, TM and the start of the sequence.
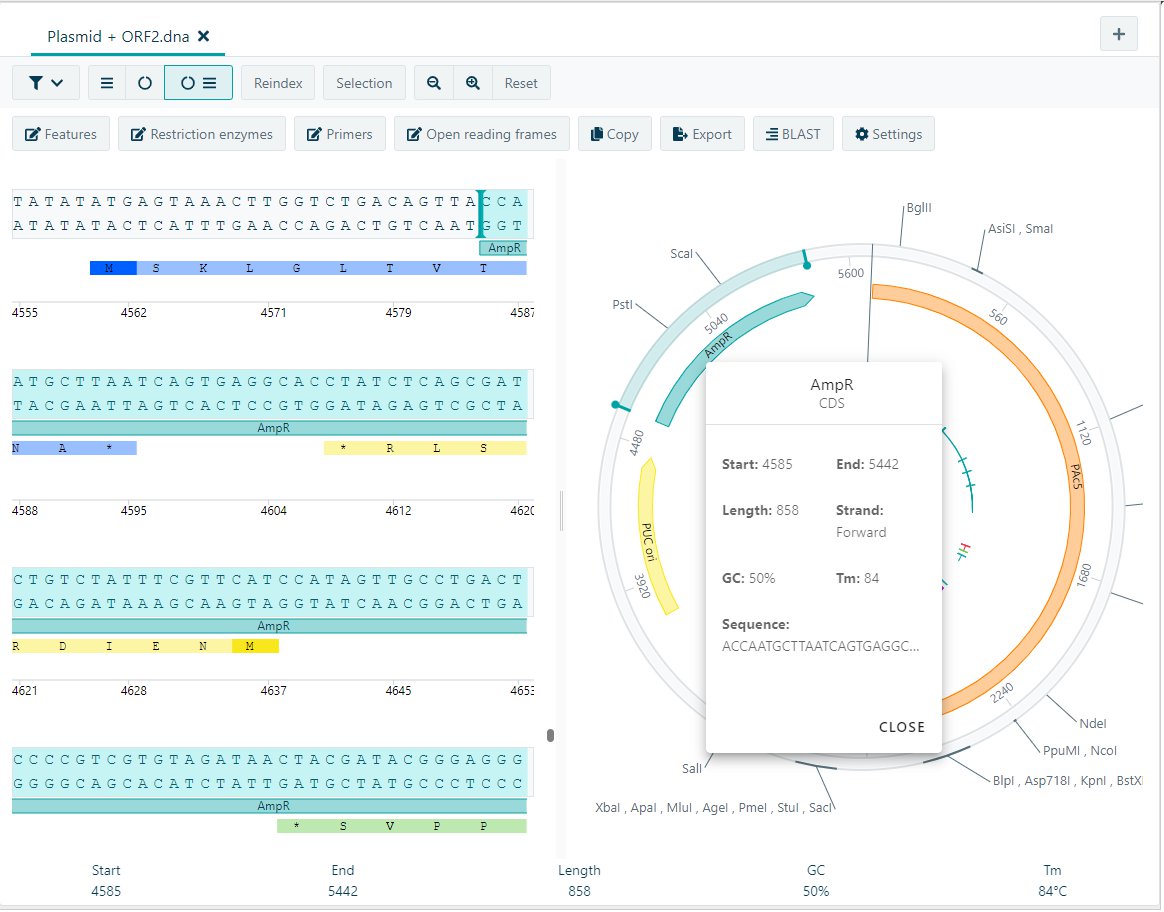
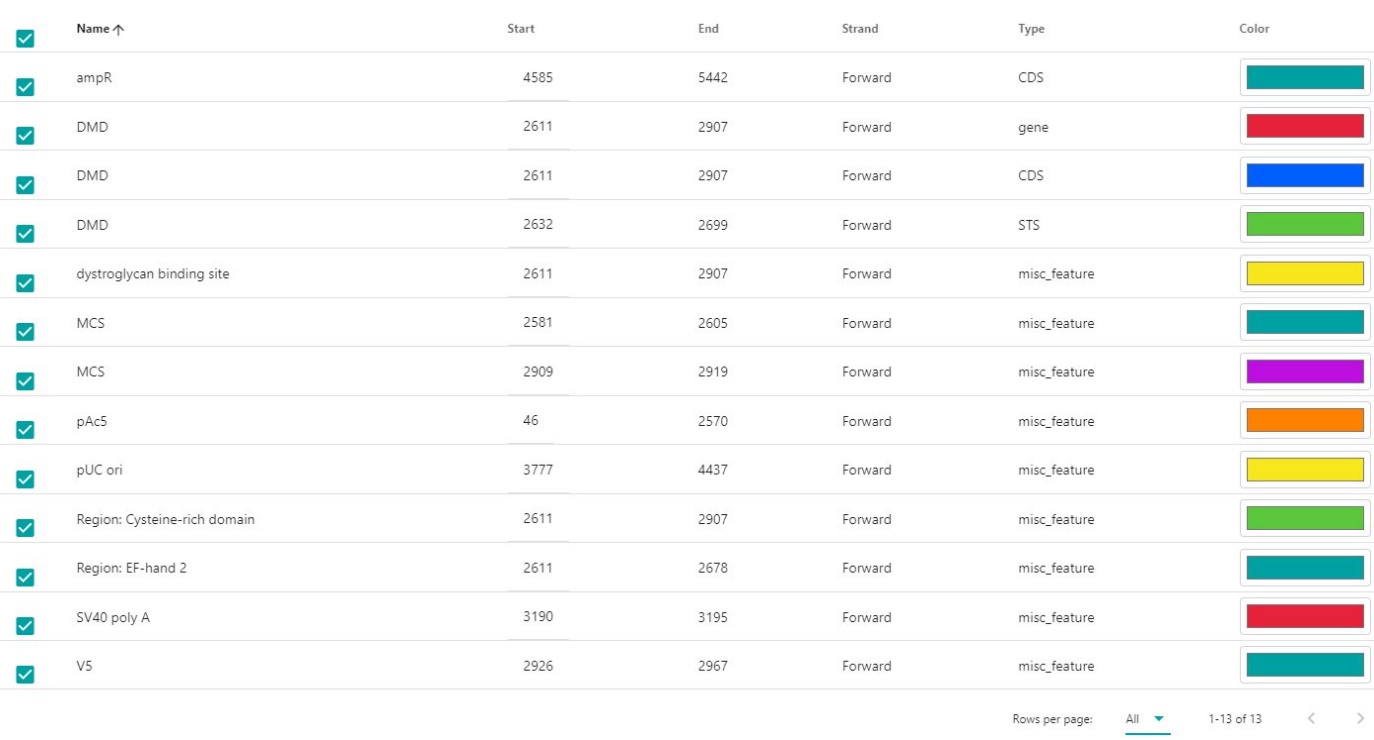
To get a list of all the features click on Features and all features available in the molecule will be displayed. If wanted the color could be changed, and data could be reorganized by clicking on Name, Start, End, Strand, or Type.
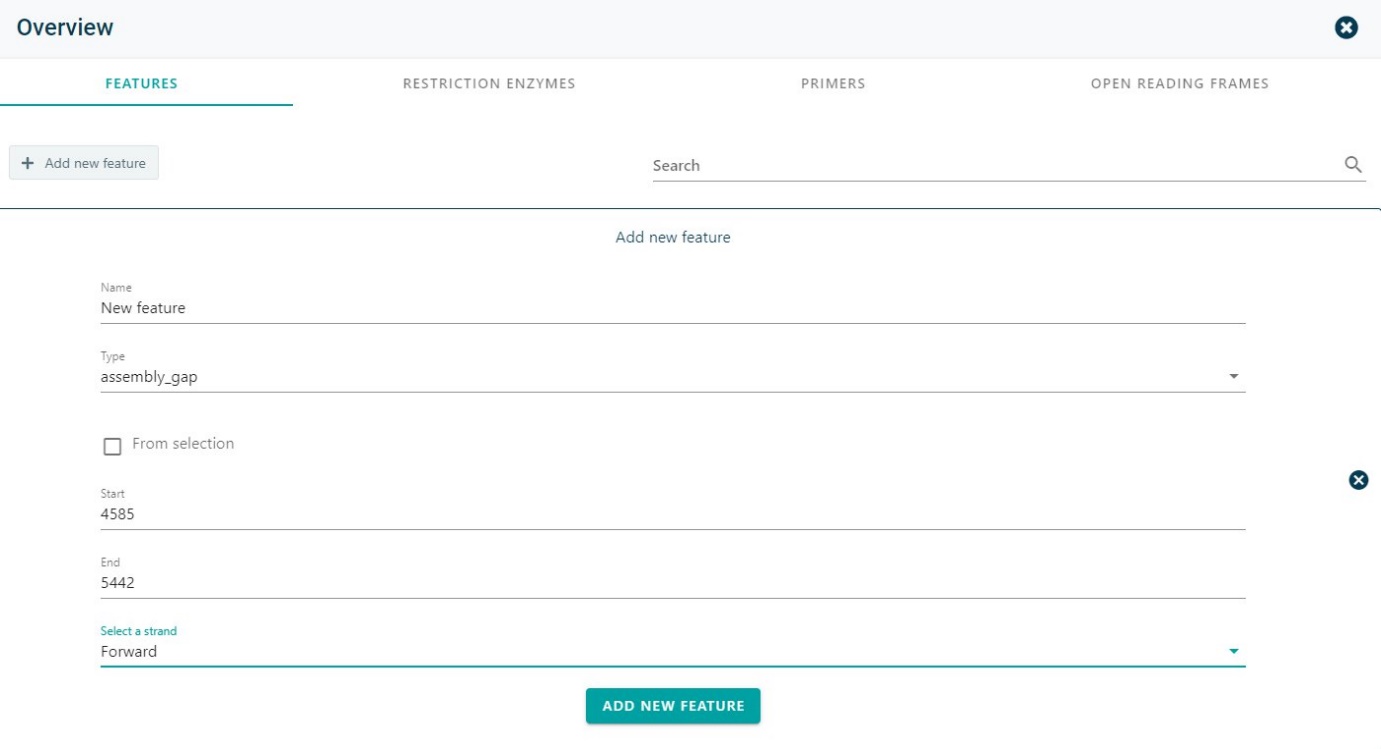
To add a new feature, click on + Add new feature. Give it a name, choose the type of feature from the dropdown menu, give the start and end, and select the strand (forward or reverse). If wanted the feature could be first selected in the sequence and this selection could be used.
Click on Add new feature and the new feature will be present in both the linear view and circular view.
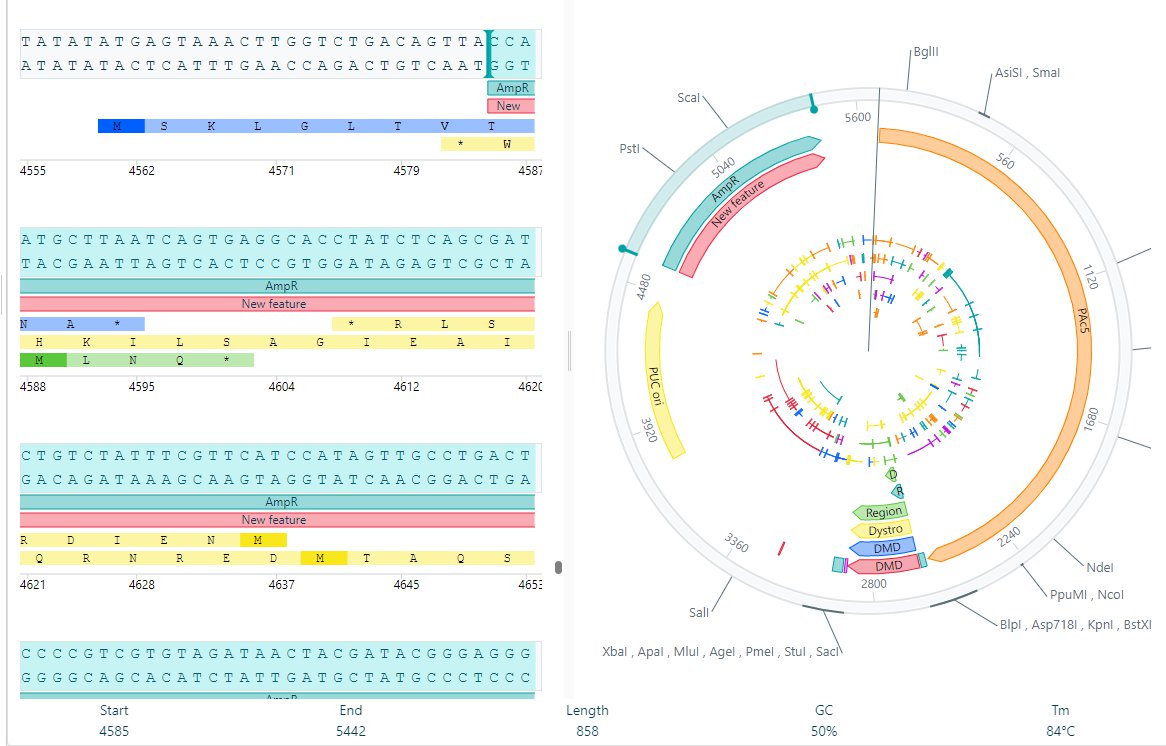
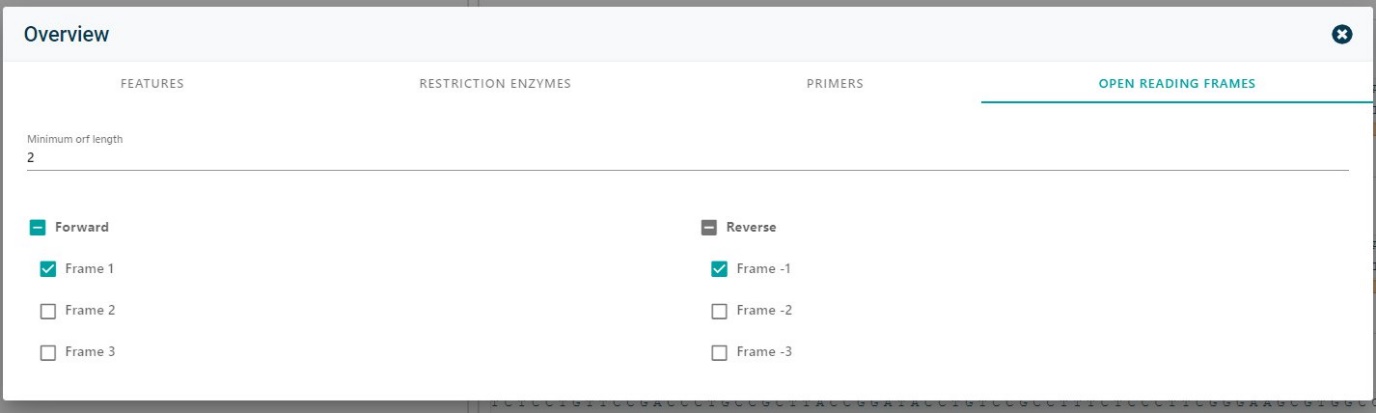
See if there are other possible open reading frames in your molecule by changing the frame. Click on Open reading frames and adjust how it is needed.
Copy your nucleotide sequence to use in any other program.
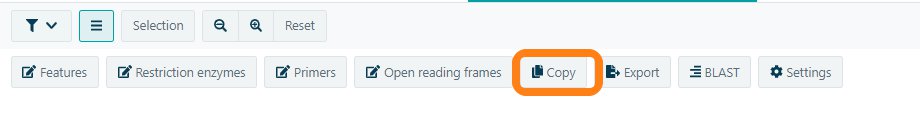
Select the desired sequence or have the complete molecule copied. Not only the nucleotide sequence can be copied, also the Amino acid translation can be copied in the multiple frames.
Export your molecule to Fast (.fasta), GenBank (.gb) or as an image by clicking on Export.
To Blast your molecule there is a direct integration with the NCBI Blast. Click on Blast and the sequence of the molecule is directly uploaded for a Blast.
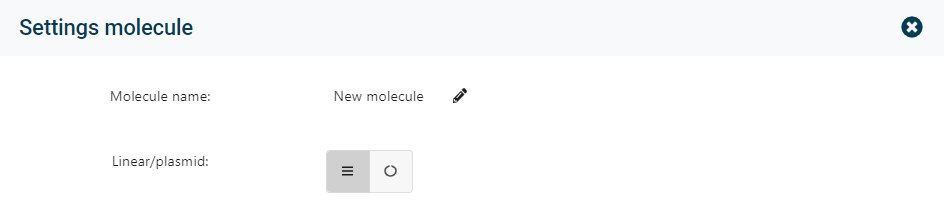
In the Settings the name of the molecule can be changed. Also the format of the molecule could be changed to Linear or Plasmid.
Digestion reactions
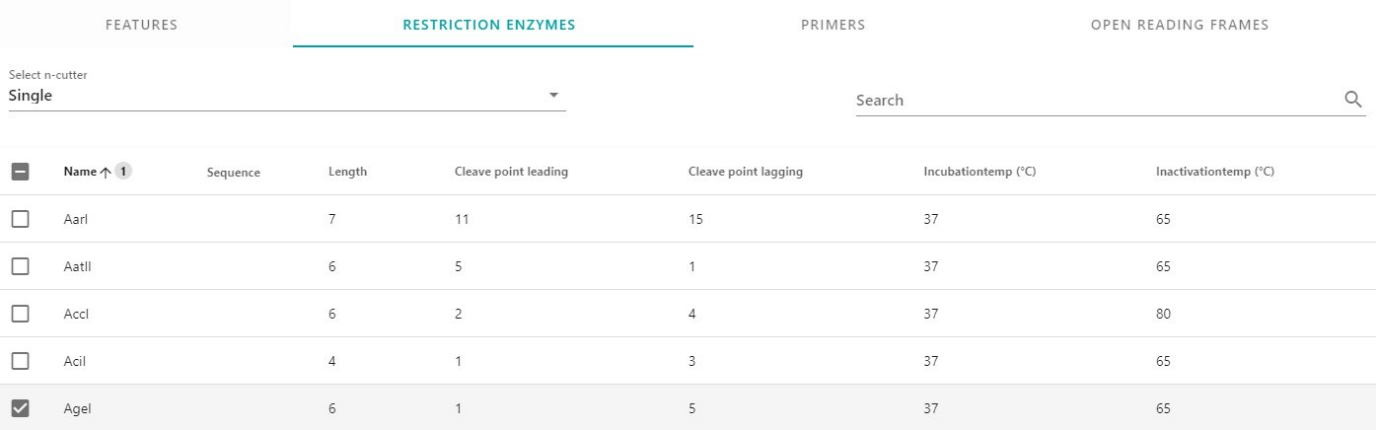
To get an overview of all the Restriction enzymes cutting in your molecule, click on Restriction enzymes. Select single, double or triple cutting enzymes via the dropdown menu or use the search option to find specific enzymes. Select the enzymes you would like to use in your digestion reaction.
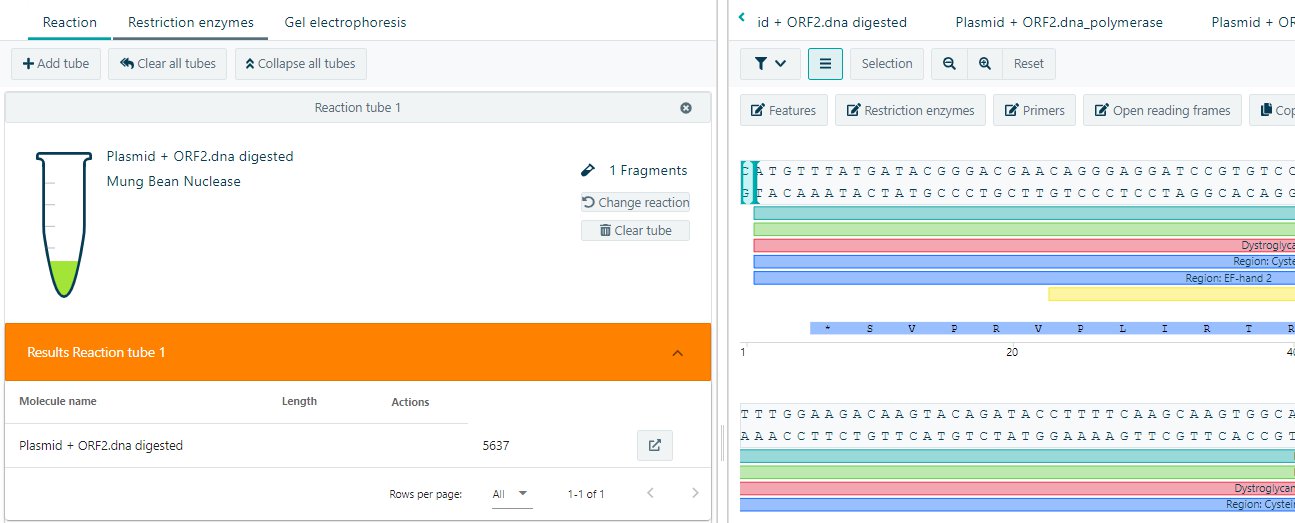
To perform a digestion reaction in CloneAssist, click on + Add Tube and a new tube will appear. Select the molecule(s) you would like to add and select the enzyme. For a digestion reaction please select restriction. Add the restriction enzyme(s) you would like to use. By clicking on Start reaction, CloneAssist will perform the reaction.
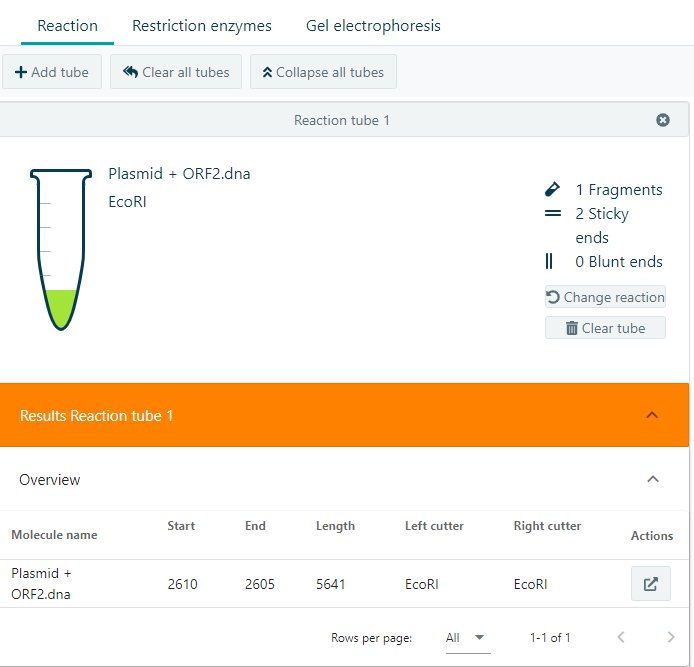
The results will show an overview of the number of fragments, number of sticky or blunt ends, and the resulting Molecule(s) including the most important information. If you would like to continue working on the resulting molecule, click on ![]() to isolate the molecule. A new molecule to work with will appear on the right side of the application.
to isolate the molecule. A new molecule to work with will appear on the right side of the application.
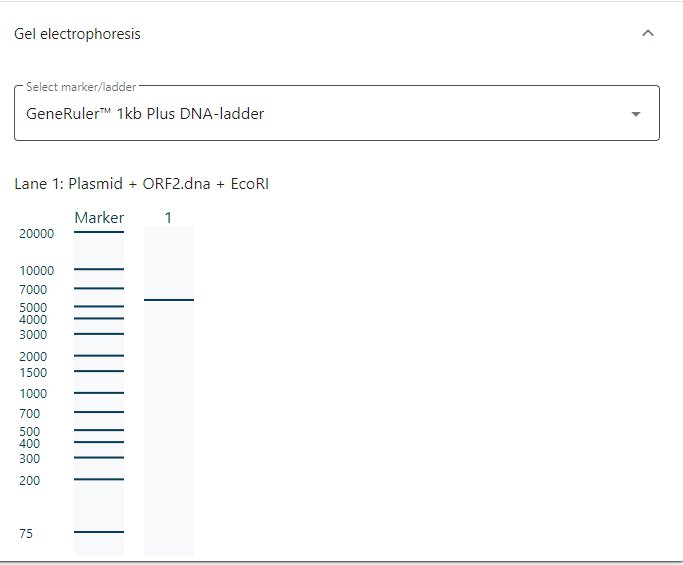
If you would like to see resulting gel electrophoresis, click on Gel electrophoresis, select you favorite marker/ladder and see how the DNA electrophoresis gel should look like.
Ligation reaction
Click on + Add tube to start the reaction. Select the molecules needed in the ligation reaction, select from the Select enzyme dropdown menu Ligase. CloneAssist will automatically show the number of possible (invalid) ligation options. Select the molecule to isolate after the reaction and start the reaction.
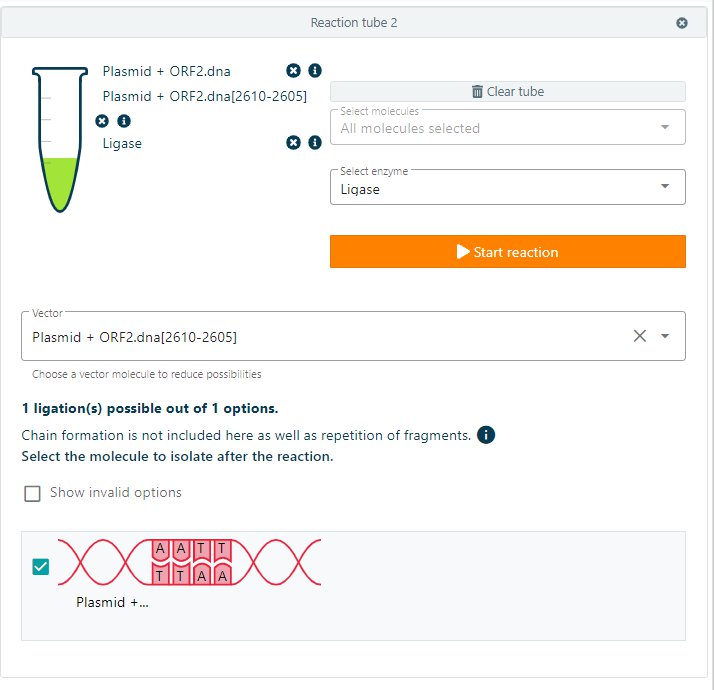
By isolating the molecule, a new molecule to work with will appear on the right side of the screen.
PCR reaction
Add primers to your molecule. Primers could be added by selecting a region or manual. Open the Primers in your molecule.
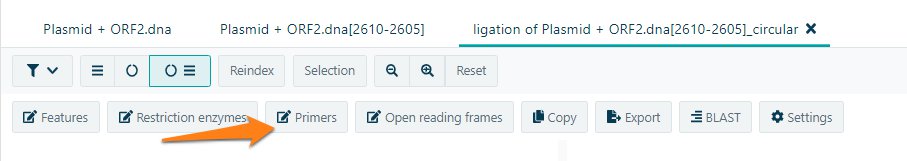
Add a new primer by selecting + Add new primer. Give the name. Select the corresponding strand from the dropdown menu. Either add your primer manual of from selection. CloneAssist wil automatically calculate Tm and GC content. Add the new primer and the primer will appear in the primer list.
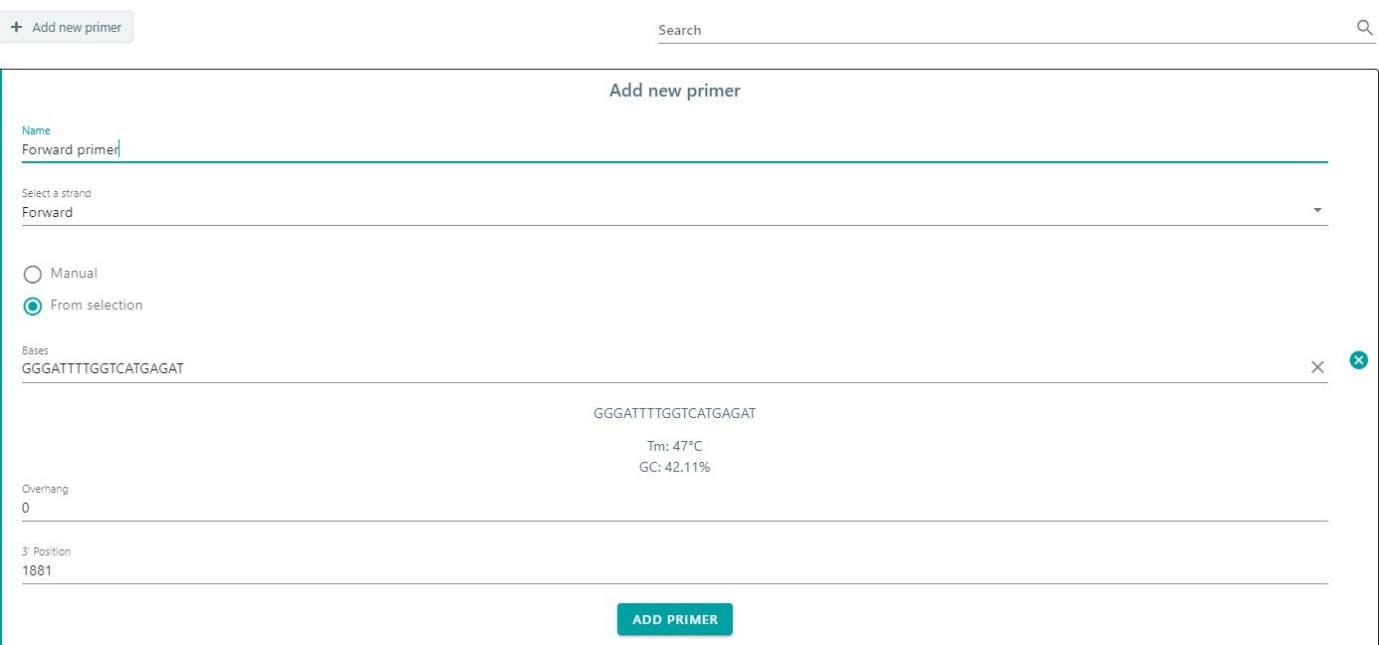
If needed the primers could be edited, deleted, or the color could be changed. In the primer list.
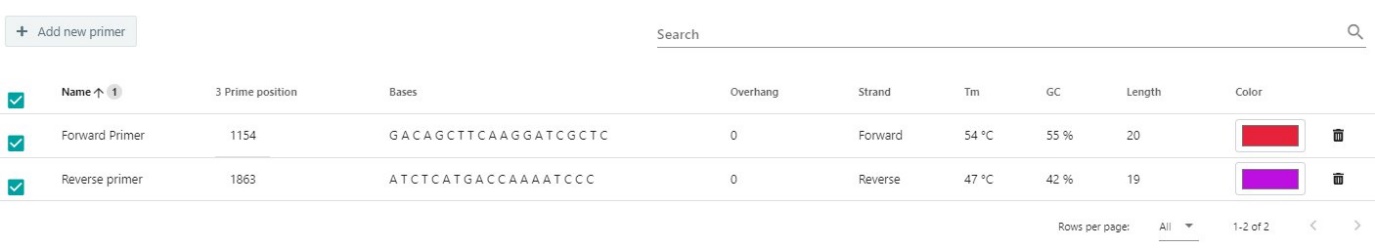
To start with the PCR reaction, add a new tube, select your molecule and select Polymerase from the select enzyme dropdown menu. Add your primers to the reaction. CloneAssist will calculate the Tm difference and the expected Product length. Start the reaction to perform the PCR reaction.
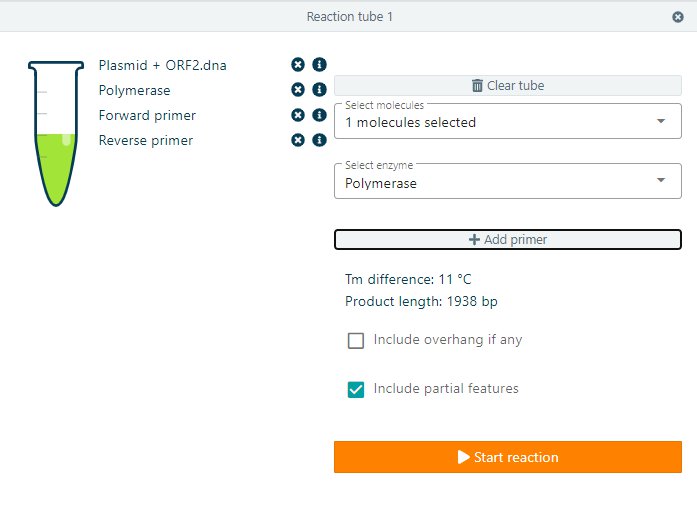
Isolate the resulting product for further analyses.
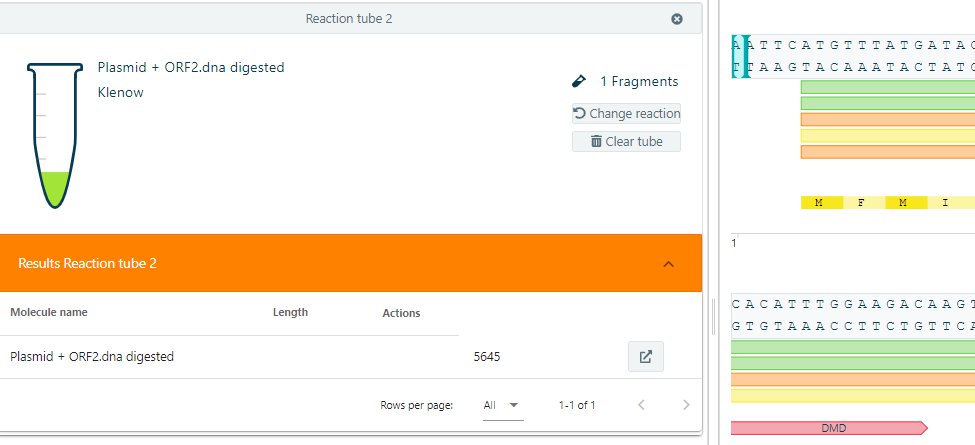
Klenow reaction
Add a new tube. Select your molecule for the Klenow reaction. Select klenow in the dropdown menu. And start the reaction. The resulting molecule can be isolated for further analysis.
Mung Bean Nuclease reaction
Add a new tube. Select the molecule for the Mung Bean Nuclease reactions. Select Mung Bean Nuclease in the dropdown menu and start the reaction. The resulting molecule can be isolated for further analysis.